This is a continued blog following Build And Compare Classification Models. We are going to build and compare a bunch of regression models in this post.
As for the program, we use the same mechanism. A factory class named RegressorFactory takes on tasks such as instantiating a model and fitting the model and predicting and evaluating the model.
Function CompareRegressionModels() takes the responsibility of implementing the work flow.
Along with the score (Coefficient of determination) provided by a model itself, root mean squared error (RMSE) is another indicator chosen to evaluate the models. You can find them on the comparison graph.
A solid circle represents a model on the chart. The score is displayed near the circle. The RMSEs of the training data and the test data stand for X, Y axis, respectively.
make_regression Dataset
Another example for California Housing dataset.
California Housing
Models to Be Compared
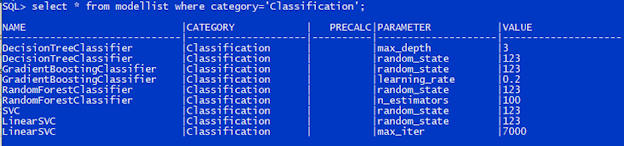
The regression models are also administered in the table ModelList with Category set to Regression, as shown on the screenshot below.
PreCalc indicator is set to 1 if the model requires polynomial calculation, otherwise set to NULL.
The pair of Parameter and Value define the parameters passed to the model in its creation. The table structure allows you to add up to unlimited parameters. What if we specified a duplicate parameter? The first one will be picked out and passed to the model.
Dataset
Dataset should be loaded into table DatasetReg. Please make sure you only add the feature columns and the label column to the table and place the label column in the last.
The sample is a dataset generated by make_regression() method.
make_regression Dataset
Main Flow
The main flow is realized in function CompareRegressionModels(), shown in the diagram below.
But one thing we need to pay attention to, some models such as Lasso, Polynomial and Ridge require transforming the input data with polynomial matrix before it is fed into the models. So, after standardize the input data, we call PolynomialFeatures() to prepare polynomial calculation matrix, then pass the standardized data to the calculation matrix and get the output. The output will be passed to that group of models. As a result, the flow becomes slightly different.
How to Add a Model?
In case you want to add more models, you can insert the corresponding records into the table ModelList using SQL scripts, or whatever database tool. And please make sure the new model has been included in class RegressorFactory. Otherwise, you will get a warning message saying the model is not implemented as of now.
insert into modellist values('Multiple', 'Regression', null, '', '');insert into modellist values('Polynomial', 'Regression', 1, '', '');insert into modellist values('Ridge', 'Regression', 1, 'alpha', '0.1');insert into modellist values('Ridge', 'Regression', 1, 'random_state', '123');
How to Switch the Dataset?
The program fetches data from table DatasetReg, so the data must be moved into DatasetReg. Here is an example for your reference.
- Create an external table named Dataset_Housing.
create table Dataset_Housing (MedInc number,HouseAge number,AveRooms number,AveBedrms number,Population number,AveOccup number,Latitude number,Longitude number,Price number)organization external(type oracle_loaderdefault directory externalfileaccess parameters(records delimited by newlinenobadfilenologfilefields terminated by ',')location ('cal_housing.csv'))reject limit unlimited;
- Drop table DatasetReg.
- Create table DatasetReg from Dataset_Housing.
GradientBoostingRegressor is used for mountain temperature prediction in the blog Machine Learning - Build A GradientBoostingRegressor Model. In effect it overfit the training dataset. So we run all these regression models on the same dataset this time.
As you can see from the evaluation graph, DecisionTreeRegressor and Polynomial and RandomForestRegressor and Ridge tend to be overfitting as well for this particular predictive case.
Reference
Python Statistics & Machine Learning Mastering Handbook Team Karupo
Machine Learning - Build A GradientBoostingRegressor Model
Machine Learning - Build And Compare Classification Models